Our Research
The Lequieu Lab uses simulation and theory to predict the structure of soft materials across molecular to microscopic length scales. The prediction of material properties across these disparate length scales is a challenge, yet is critically important for the design of new materials with tailored properties. In the Lequieu Lab, we are interested in problems where hierarchical interactions are thought to be important, yet where a fundamental understanding of how these interactions control structure is absent.
Fast and accurate multi-scale models
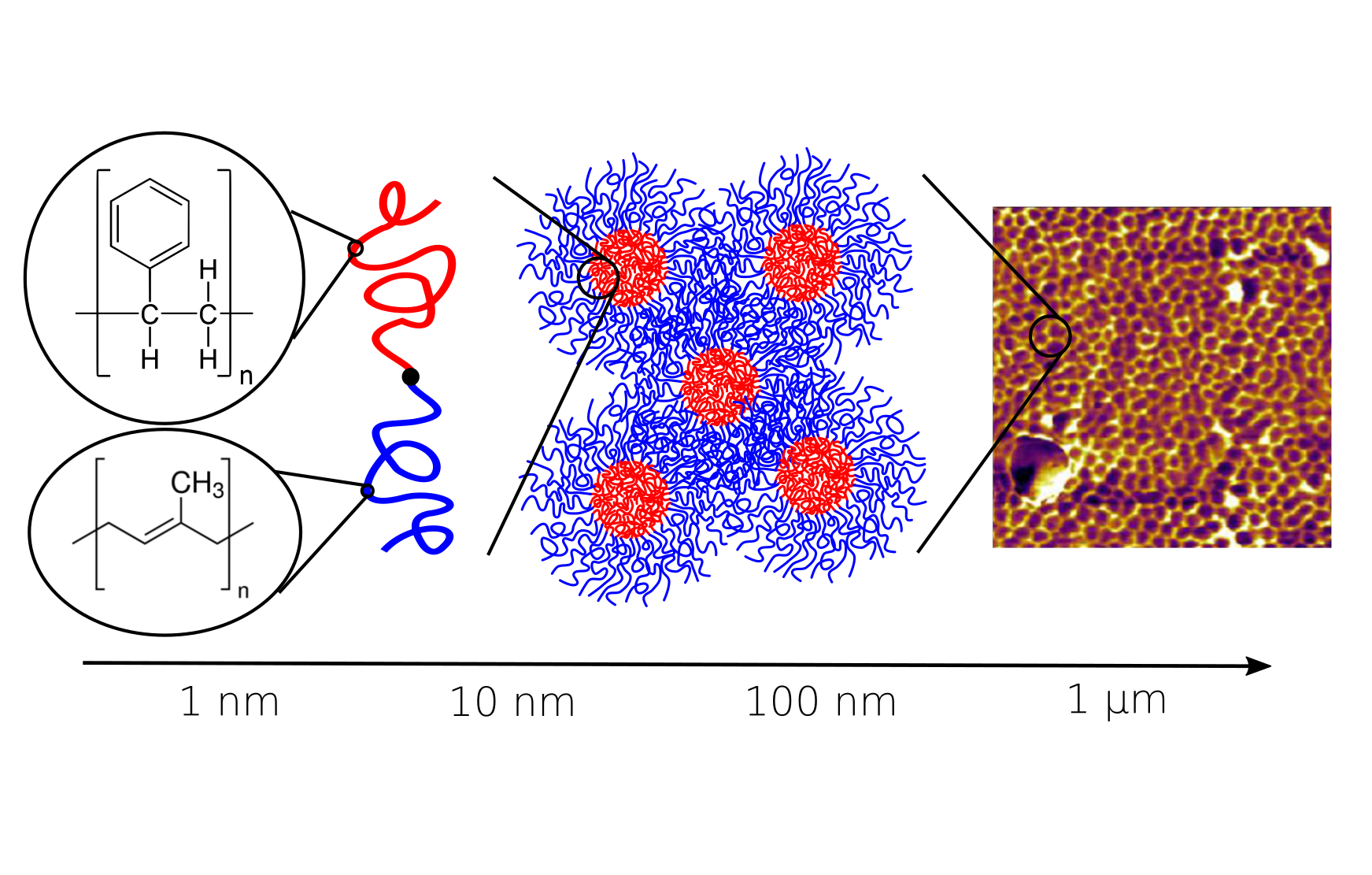
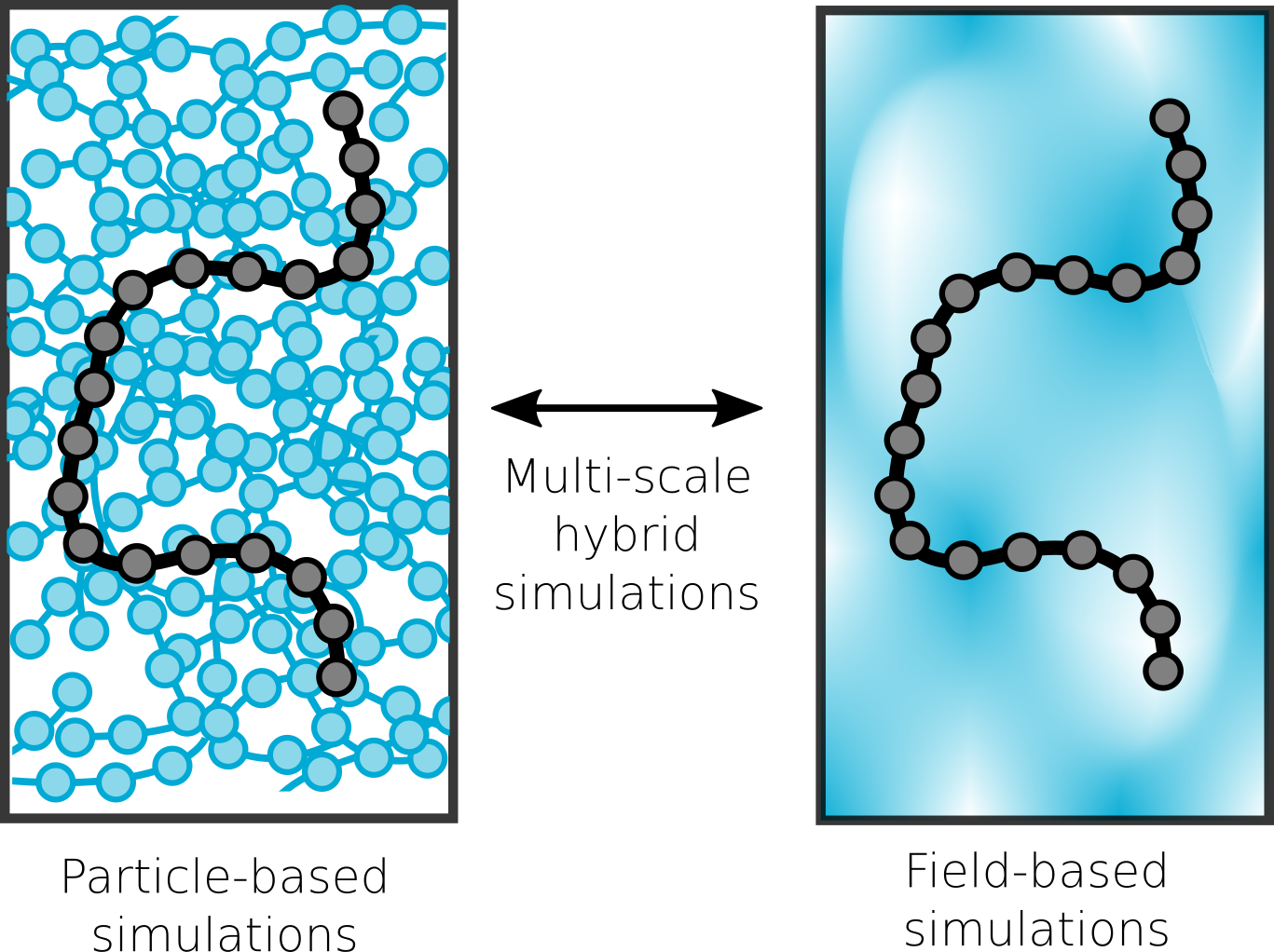
A central focus of the group the accurate prediction of structure in polymeric materials. The challenges associated with multi-scale materials are particularly acute in polymers, where the wide range of relaxation processes make it challenging to relate subtle monomer-level interactions to mesoscopic assemblies. To surmount these challenges, we are developing novel simulation techniques that combine field-theoretic and molecular dynamics simulations into a single unified framework. These approaches combine the advantages of both methodologies to rapidly equilibrate large molecular-weight polymer melts, while still maintaining access to molecular configurations and chain dynamics.
New paradigms for self-assembly
Another topic of interest is the development of new approaches that can expand the palette of possible structures that can be achieved with block copolymers. Despite the widespread applications block copolymers in society, these extremely versatile molecules only self-assemble into a relatively small set of microphases. New applications areas ranging from water filtration to advanced optics will necessitate the expansion of this possible set of phases.

Biological polymers
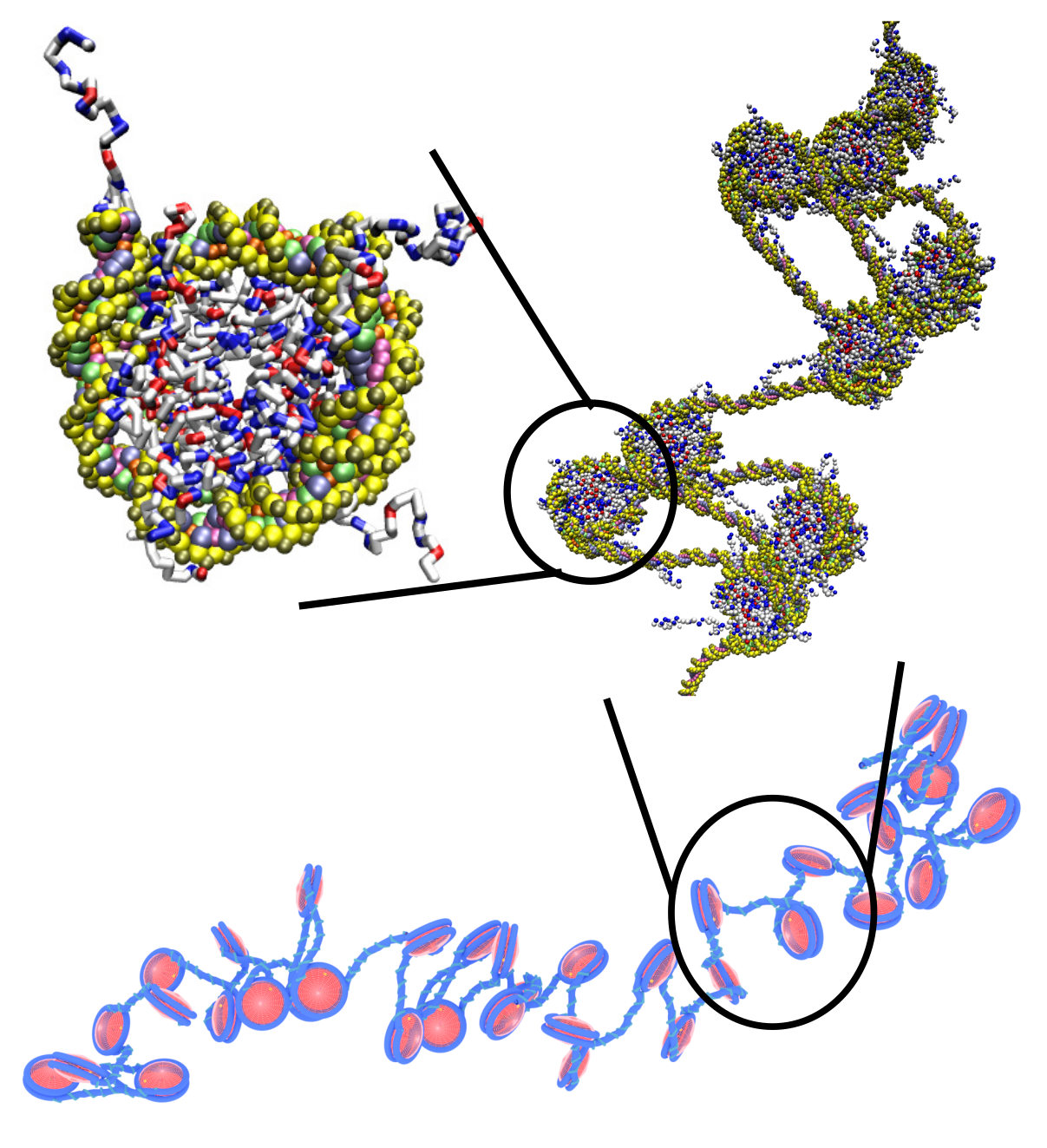
A long-time interest of the group is to understand the self-assembly of biological polymers, especially DNA and chromatin. We have developed several widely-used models of chromatin that can be used to examine length scales ranging from a single nucleosome to many kilo-bases of DNA. We are using these tools to understand the relationship between histone modifications and chromatin's 3-dimensional structure.
