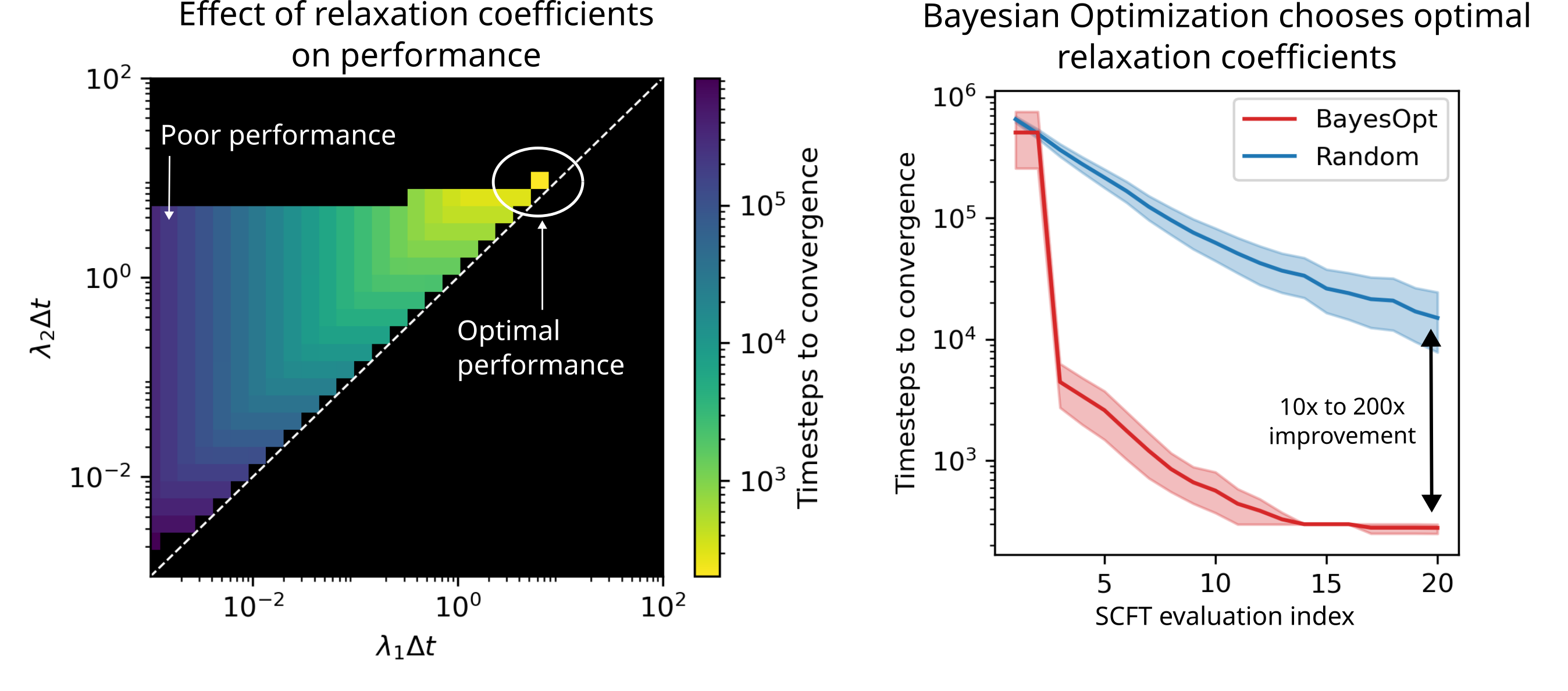
Accelerating Multi-Species Field-Theoretic Simulations Using Bayesian Optimization
Ritvind Suketana, Andrew Golembeski, Joshua Lequieu
Molecular Systems Design & Engineering (2025) 10:982--996 DOI: 10.1039/D5ME00100E
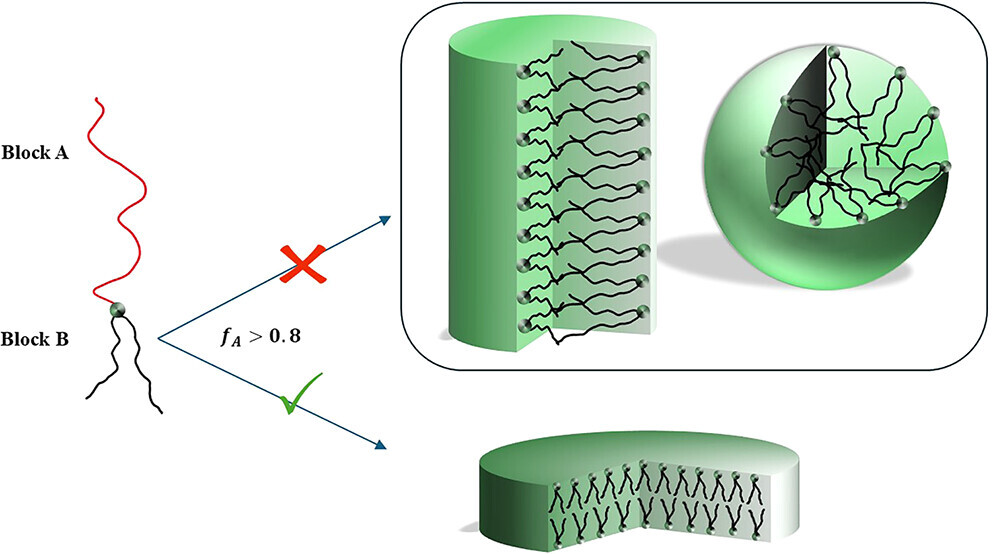
Morphological Transitions and Chain Conformations in AB2 Miktoarm Star Block Copolymers: A Molecular Dynamics Study
Zerihun G. Workineh, Farzad Toiserkani, Joshua Lequieu, Giuseppe Pellicane, Mesfin Tsige
Macromolecules (2025) 58:3343--3354 DOI: 10.1021/acs.macromol.5c00029
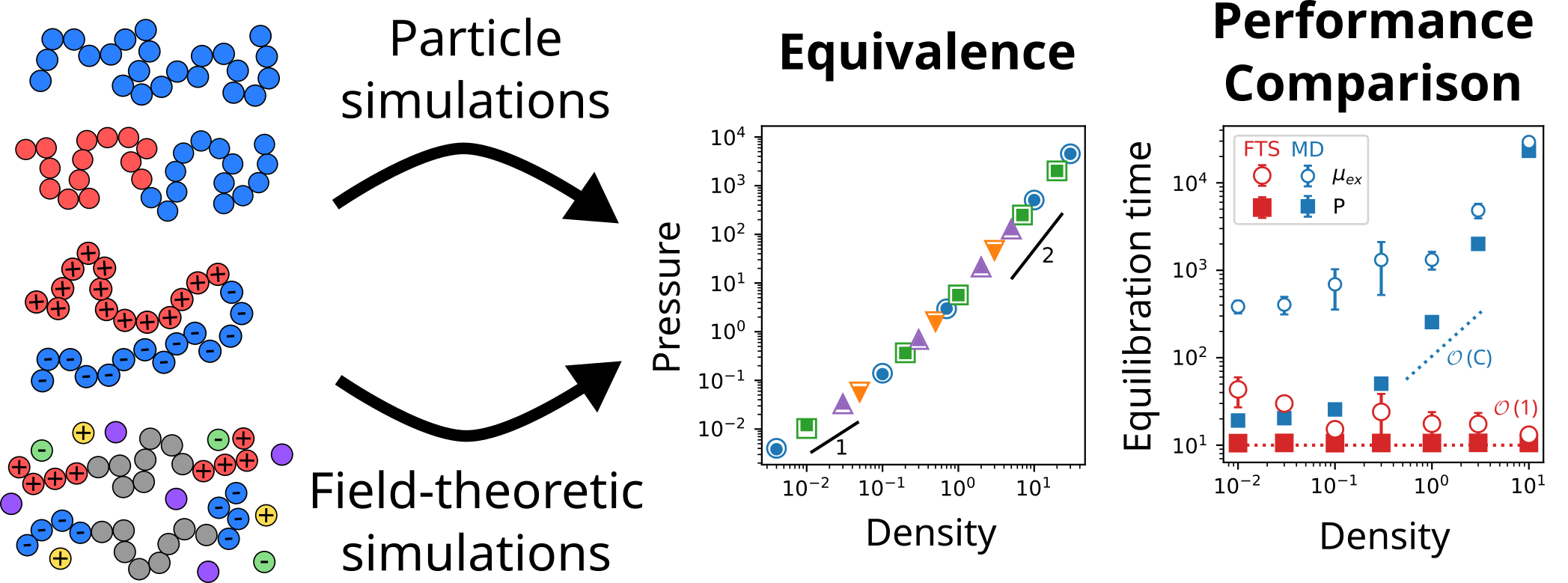
Quantitative Equivalence and Performance Comparison of Particle and Field-Theoretic Simulations
Joshua Lequieu
Macromolecules (2024) 57:10870--10884 DOI: 10.1021/acs.macromol.4c02034
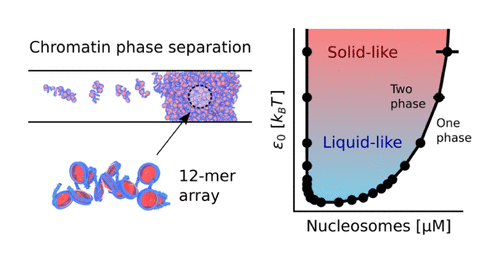
A Molecular View into the Structure and Dynamics of Phase-Separated Chromatin
Andrew Golembeski, Joshua Lequieu
The Journal of Physical Chemistry B (2024) 128:10593--10603 DOI: 10.1021/acs.jpcb.4c04420
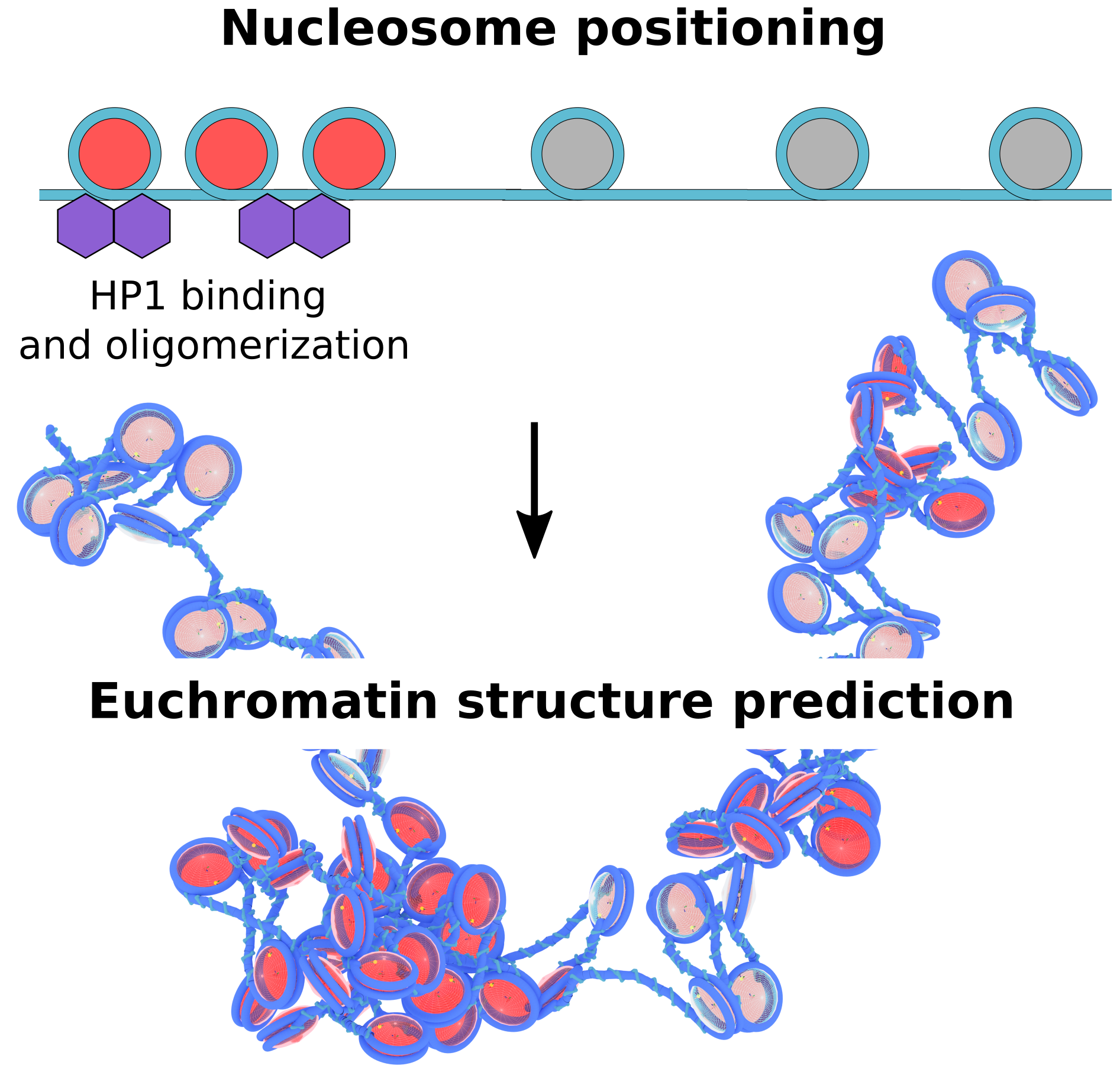
A Physical Model of Euchromatin Organization
Joshua Lequieu
Proc. Nat. Acad. Sci. USA. (2024) 121:e2410751121 DOI: 10.1073/pnas.2410751121
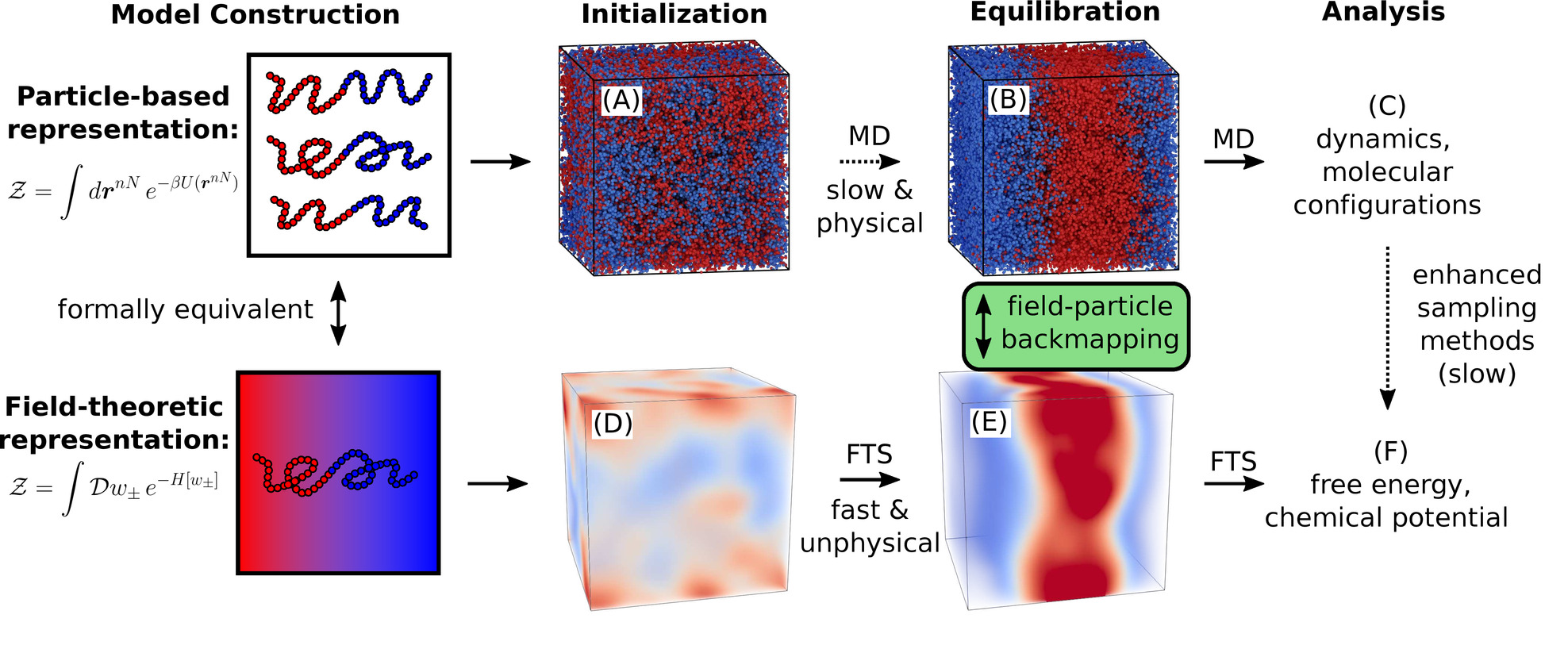
Combining Particle and Field-Theoretic Polymer Models with Multi-Representation Simulations
Joshua Lequieu
J. Chem. Phys. (2023) 158:244902 DOI: 10.1063/5.0153104
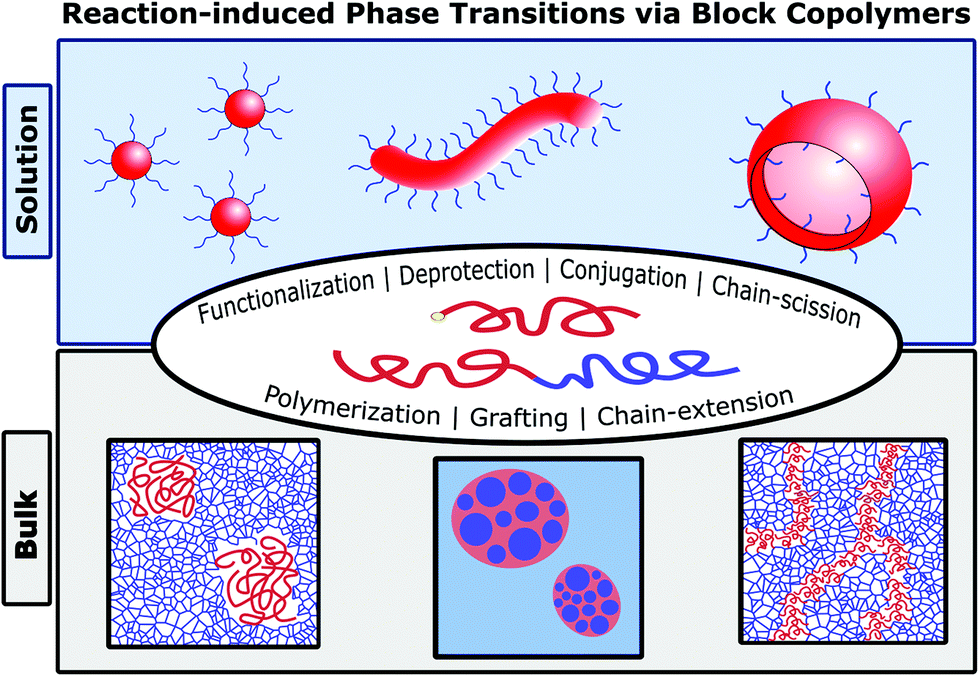
Reaction-induced phase transitions with block copolymers in solution and bulk
Joshua Lequieu, Andrew J. D. Magenau
Polym. Chem. (2021) 12:12--28 DOI: 10.1039/d0py00722f
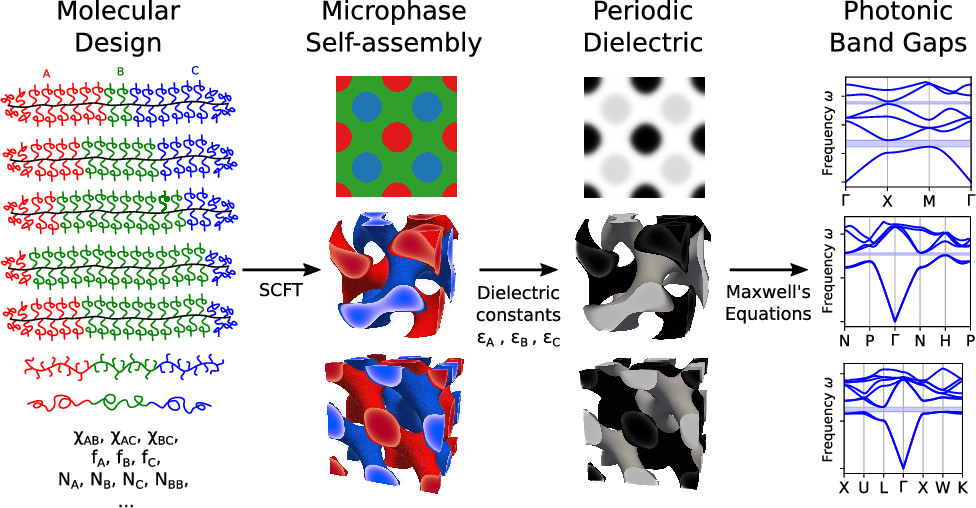
Complete Photonic Band Gaps with Nonfrustrated ABC Bottlebrush Block Polymers
Joshua Lequieu, Timothy Quah, Kris T Delaney, Glenn H Fredrickson
ACS Macro Lett. (2020) 9:1074--1080 DOI: 10.1021/acsmacrolett.0c00380
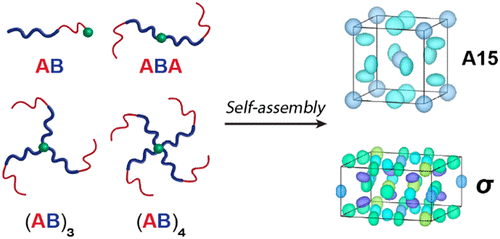
Architecture Effects in Complex Spherical Assemblies of (AB)n-Type Block Copolymers
Stephanie M. Barbon, Jung-Ah Song, Duyu Chen, Cheng Zhang, Joshua Lequieu, Kris T. Delaney, Athina Anastasaki, Manon Rolland, Glenn H. Fredrickson, Morgan W. Bates, Craig J. Hawker, Christopher M. Bates
ACS Macro Lett. (2020) 9:1745-1752 DOI: 10.1021/acsmacrolett.0c00704
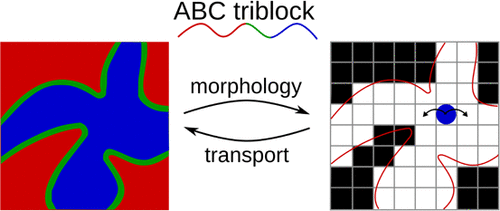
Connecting Solute Diffusion to Morphology in Triblock Copolymer Membranes
Michael P. Howard*, Joshua Lequieu*, Kris T. Delaney, Venkat Ganesan, Glenn H. Fredrickson, Thomas M. Truskett *equal contribution
Macromolecules (2020) 53:2336--2343 DOI: 10.1021/acs.macromol.0c00104
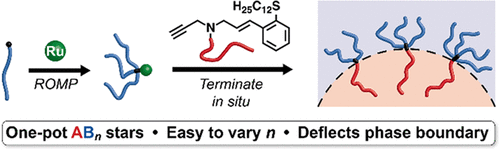
Efficient Synthesis of Asymmetric Miktoarm Star Polymers
Adam E Levi, Liangbing Fu, Joshua Lequieu, Jacob D Horne, Jacob Blankenship, Sanjoy Mukherjee, Tianqi Zhang, Glenn H Fredrickson, Will R Gutekunst, Christopher M Bates
Macromolecules (2020) 53:702--710 DOI: 10.1021/acs.macromol.9b02380
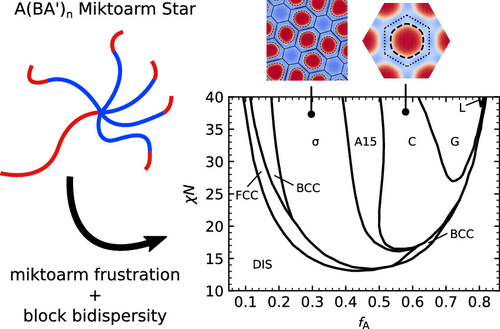
Extreme Deflection of Phase Boundaries and Chain Bridging in A(BA)́ n Miktoarm Star Polymers
Joshua Lequieu, Trenton Koeper, Kris T Delaney, Glenn H Fredrickson
Macromolecules (2020) 53:513--522 DOI: 10.1021/acs.macromol.9b02254
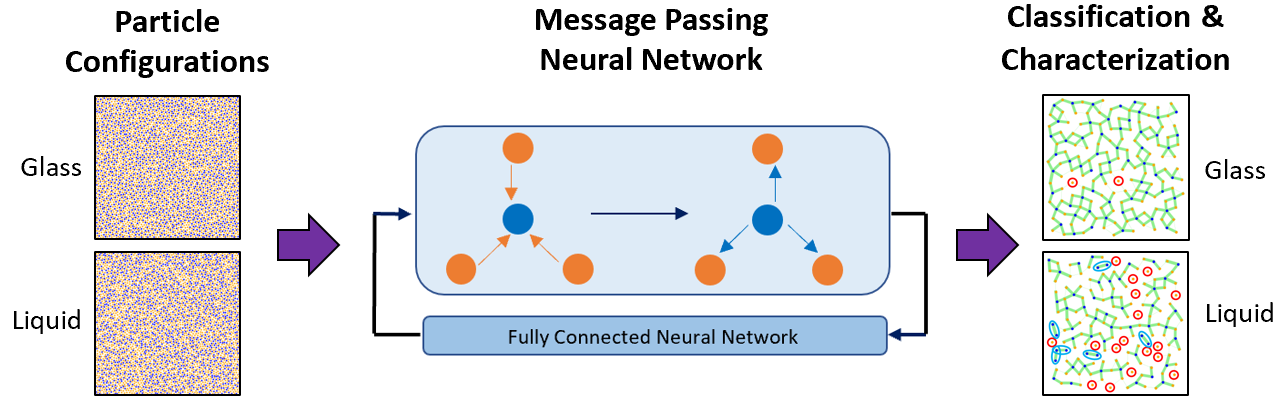
Deep Learning for Automated Classification and Characterization of Amorphous Materials
Kirk Swanson, Shubhendu Trivedi, Joshua Lequieu, Kyle Swanson, Risi Kondor
Soft Matter (2020) 16:435--446 DOI: 10.1039/c9sm01903k
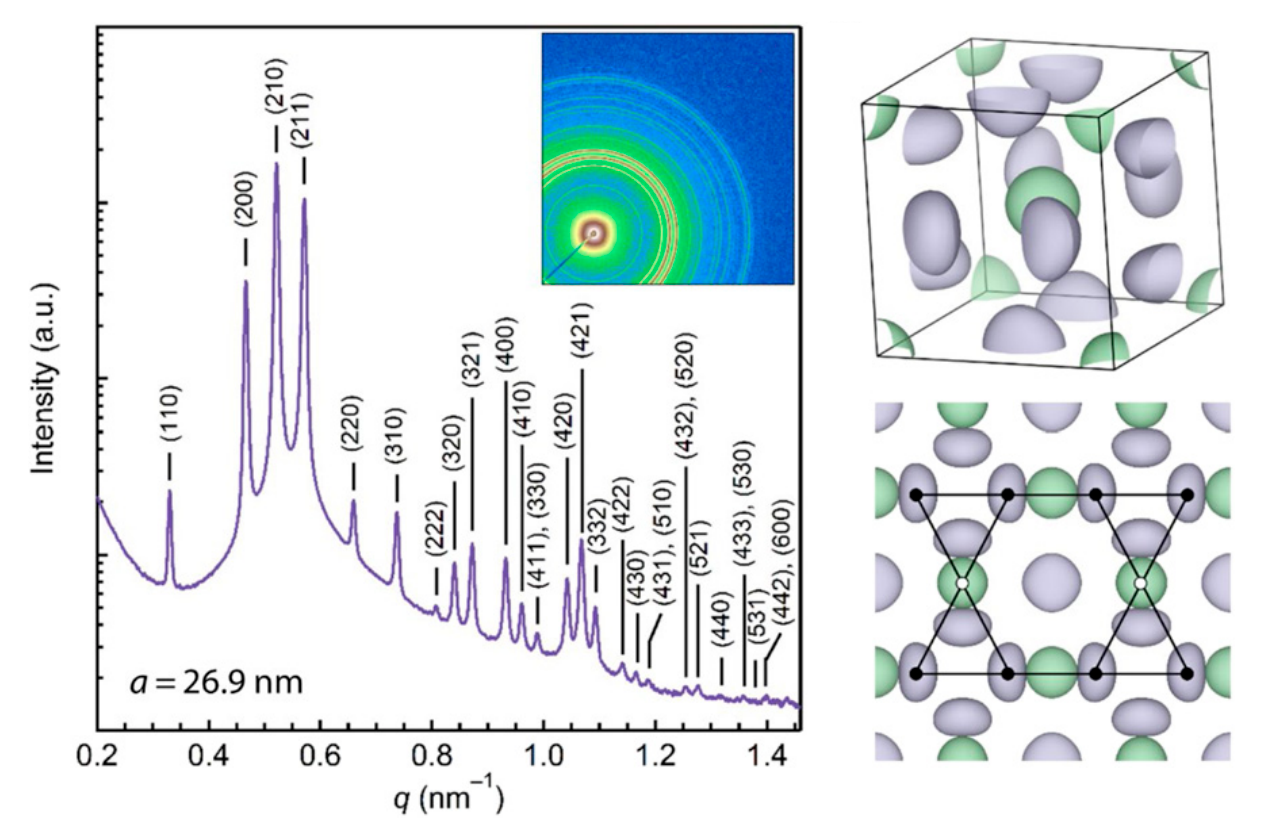
Stability of the A15 phase in diblock copolymer melts
Morgan W. Bates*, Joshua Lequieu*, Stephanie M. Barbon, Ronald M. Lewis, Kris T. Delaney, Athina Anastasaki, Craig J Hawker, Glenn H. Fredrickson, Christopher M. Bates *equal contribution
Proc. Nat. Acad. Sci. USA. (2019) 116:13194--13199 DOI: 10.1073/pnas.1900121116
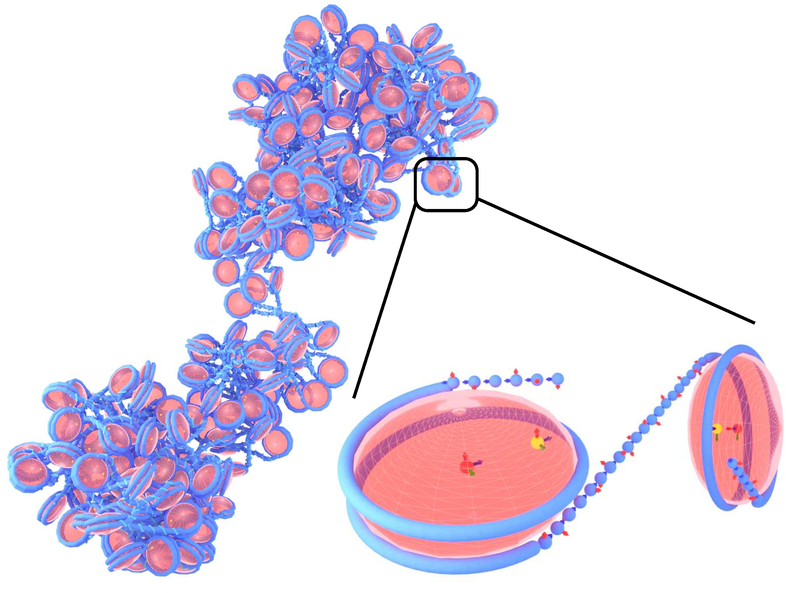
1CPN : A coarse-grained multi-scale model of chromatin
Joshua Lequieu, Andrés Córdoba, Joshua Moller, Juan J de Pablo
J. Chem. Phys. (2019) 150:215102 DOI: 10.1063/1.5092976
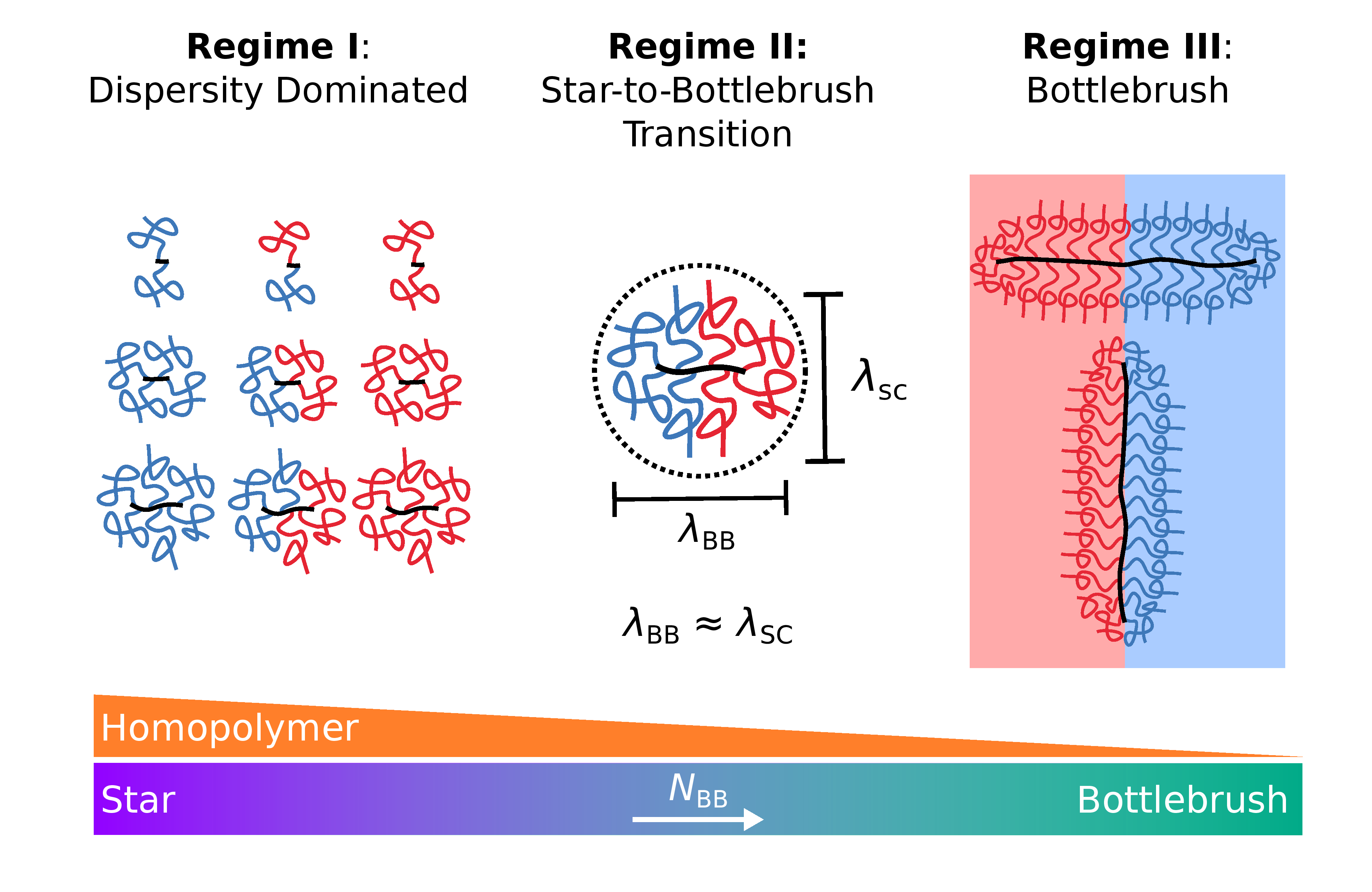
Miktoarm Stars via Grafting-Through Copolymerization: Self-Assembly and the Star-to-Bottlebrush Transition
Adam E. Levi*, Joshua Lequieu*, Jacob D. Horne, Morgan W. Bates, Jing M. Ren, Kris T. Delaney, Glenn H. Fredrickson, Christopher M. Bates *equal contribution
Macromolecules (2019) 52:1794--1802 DOI: 10.1021/acs.macromol.8b02321
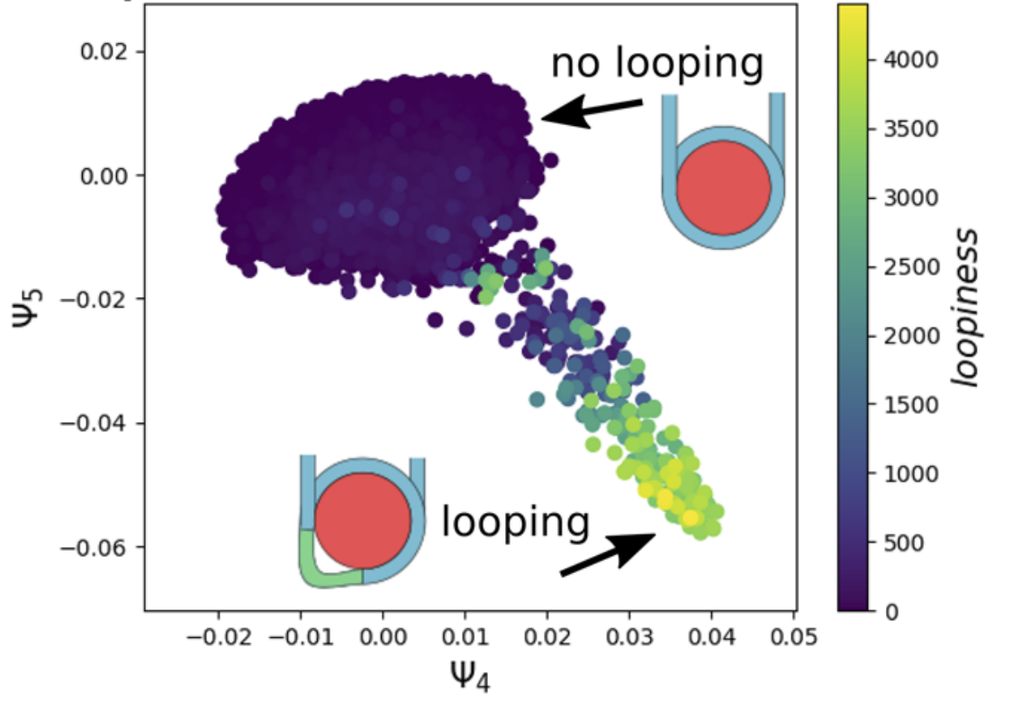
Extracting collective motions underlying nucleosome dynamics via nonlinear manifold learning
Ashley Z. Guo, Joshua Lequieu, Juan J. de Pablo
J. Chem. Phys. (2019) 150:054902 DOI: 10.1063/1.5063851
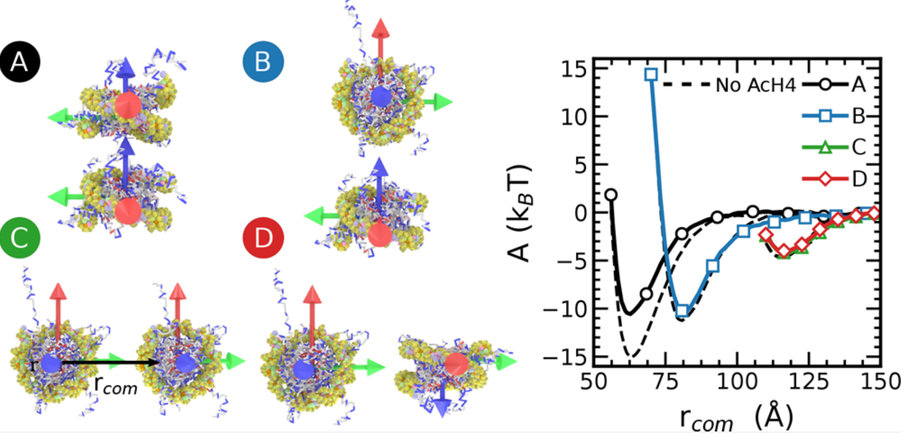
The Free Energy Landscape of Internucleosome Interactions and Its Relation to Chromatin Fiber Structure
Joshua Moller, Joshua Lequieu, Juan J. de Pablo
ACS Cent. Sci. (2019) 5:341--348 DOI: 10.1021/acscentsci.8b00836

SSAGES: Software Suite for Advanced General Ensemble Simulations
Hythem Sidky*, Yamil J. Colón*, Julian Helfferich, Benjamin J. Sikora, Cody Bezik, Weiwei Chu, Federico Giberti, Ashley Z. Guo, Xikai Jiang, Joshua Lequieu, Jiyuan Li, Joshua Moller, Michael J. Quevillon, Mohammad Rahimi, Hadi Ramezani-Dakhel, Vikramjit S. Rathee, Daniel R. Reid, Emre Sevgen, Vikram Thapar, Michael A. Webb, Jonathan K. Whitmer, Juan J. de Pablo *equal contribution
J. Chem. Phys. (2018) 148:18500 DOI: 10.1063/1.5008853
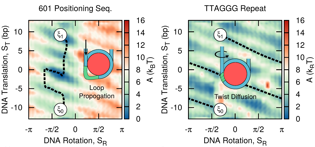
In silico evidence for sequence-dependent nucleosome sliding
Joshua Lequieu, David C. Schwartz, Juan J. de Pablo
Proc. Nat. Acad. Sci. USA. (2017) 114:E9197--E9205 DOI: 10.1073/pnas.1705685114
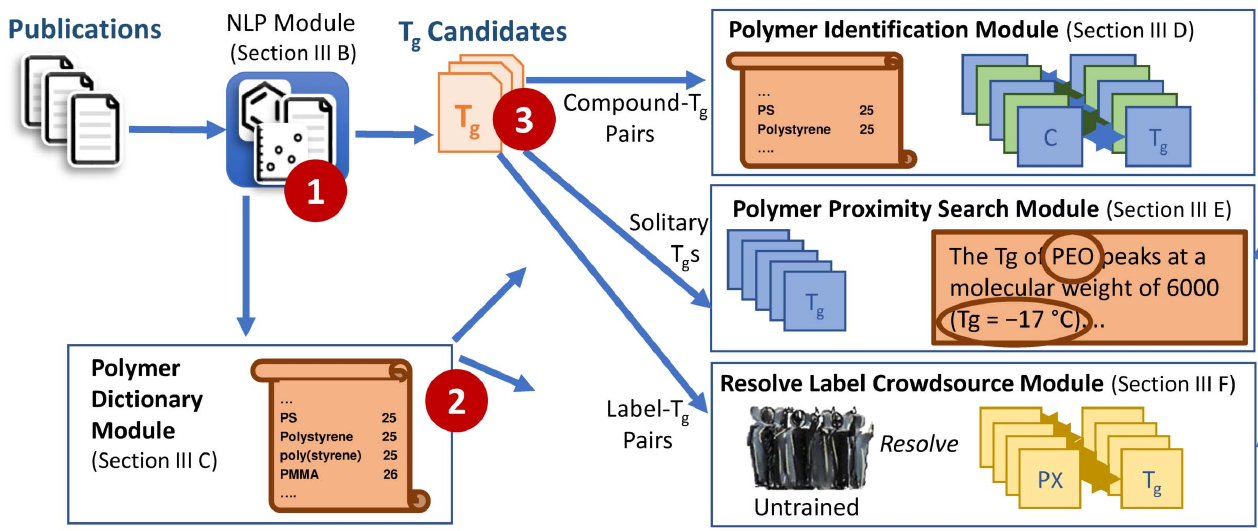
Towards a Hybrid Human-Computer Scientific Information Extraction Pipeline
Roselyne B. Tchoua, Kyle Chard, Debra J. Audus, Logan T. Ward, Joshua Lequieu, Juan J. de Pablo, Ian T. Foster
2017 IEEE 13th International Conference on eScience (2017) :109--118 DOI: 10.1109/eScience.2017.23
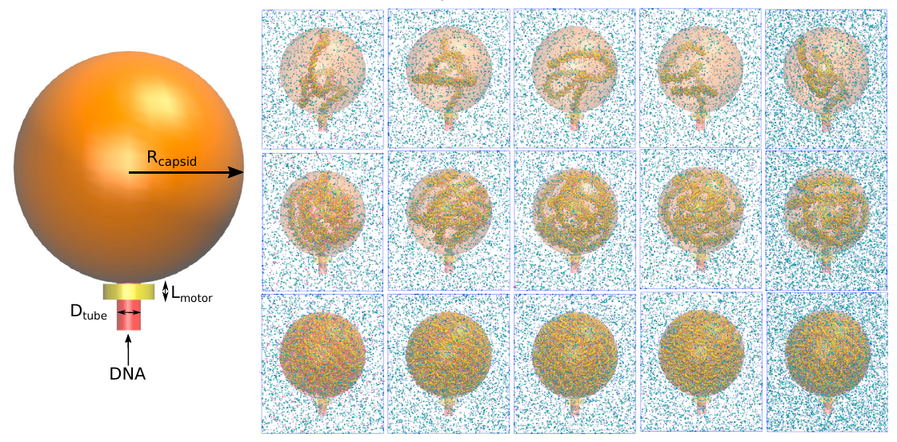
A Molecular View of the Dynamics of dsDNA Packing Inside Viral Capsids in the Presence of Ions
Andrés Córdoba, Daniel M. Hinckley, Joshua Lequieu, Juan J. de Pablo
Biophys. J. (2017) 112:1302--1315 DOI: 10.1016/j.bpj.2017.02.015
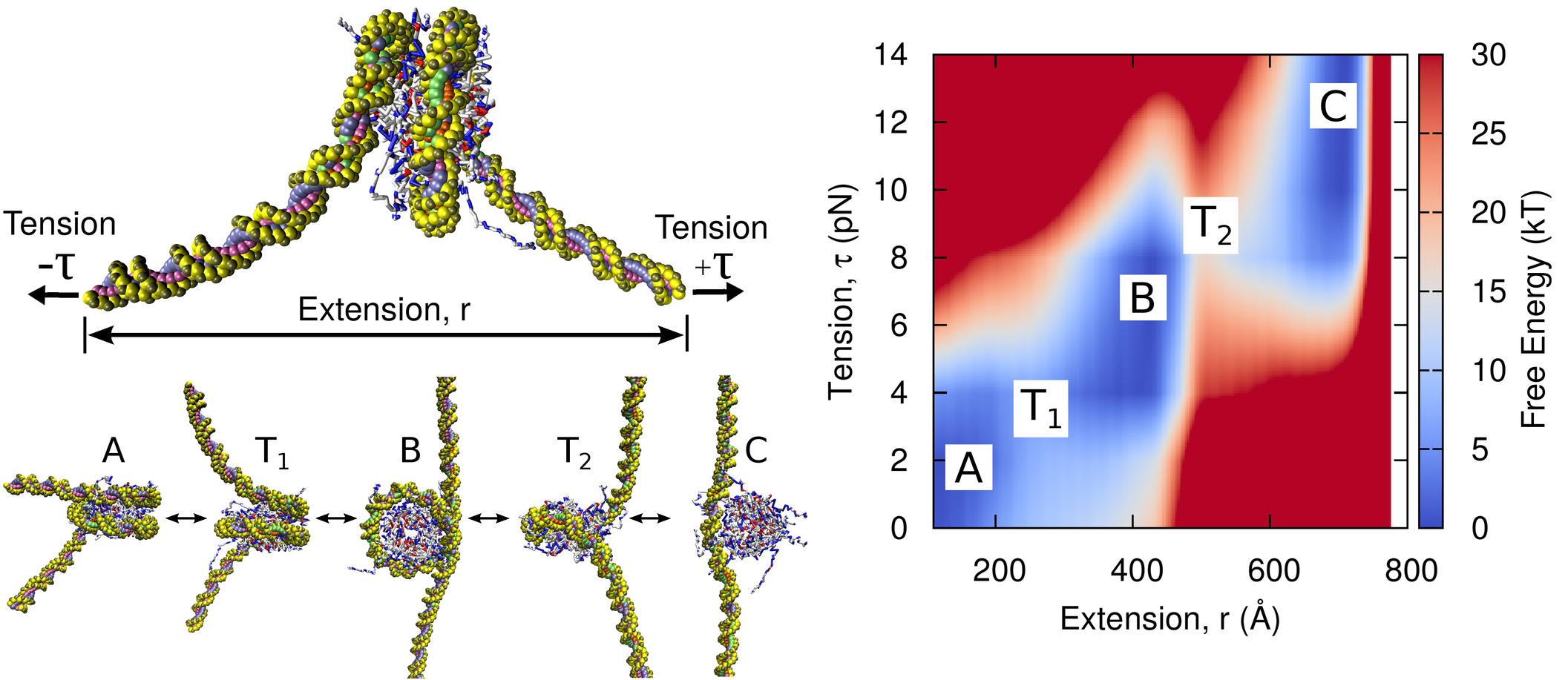
Tension-dependent free energies of nucleosome unwrapping
Joshua Lequieu, Andrés Córdoba, David C. Schwartz, Juan J. de Pablo
ACS Cent. Sci. (2016) 2:660--666 DOI: 10.1021/acscentsci.6b00201
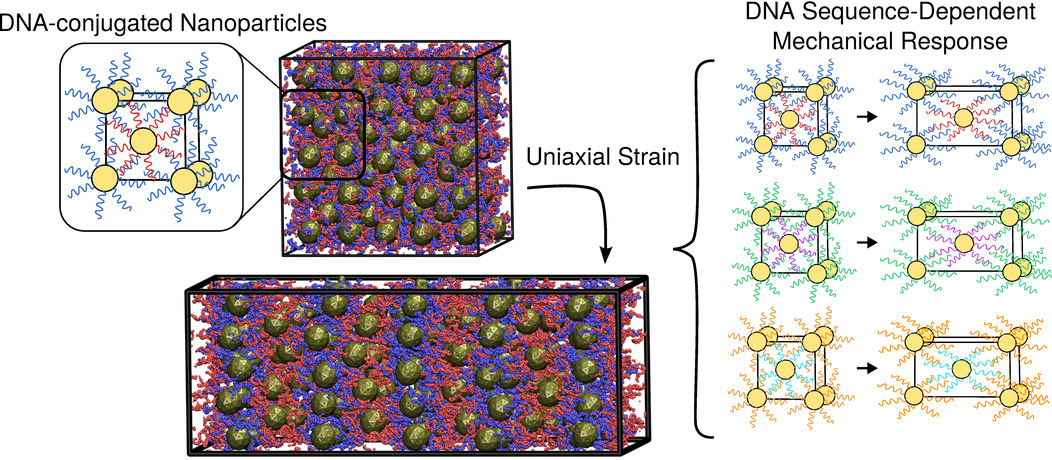
Mechanical response of DNA-nanoparticle crystals to controlled deformation
Joshua Lequieu, Andrés Córdoba, Daniel Hinckley, Juan J. de Pablo
ACS Cent. Sci. (2016) 2:614--620 DOI: 10.1021/acscentsci.6b00170
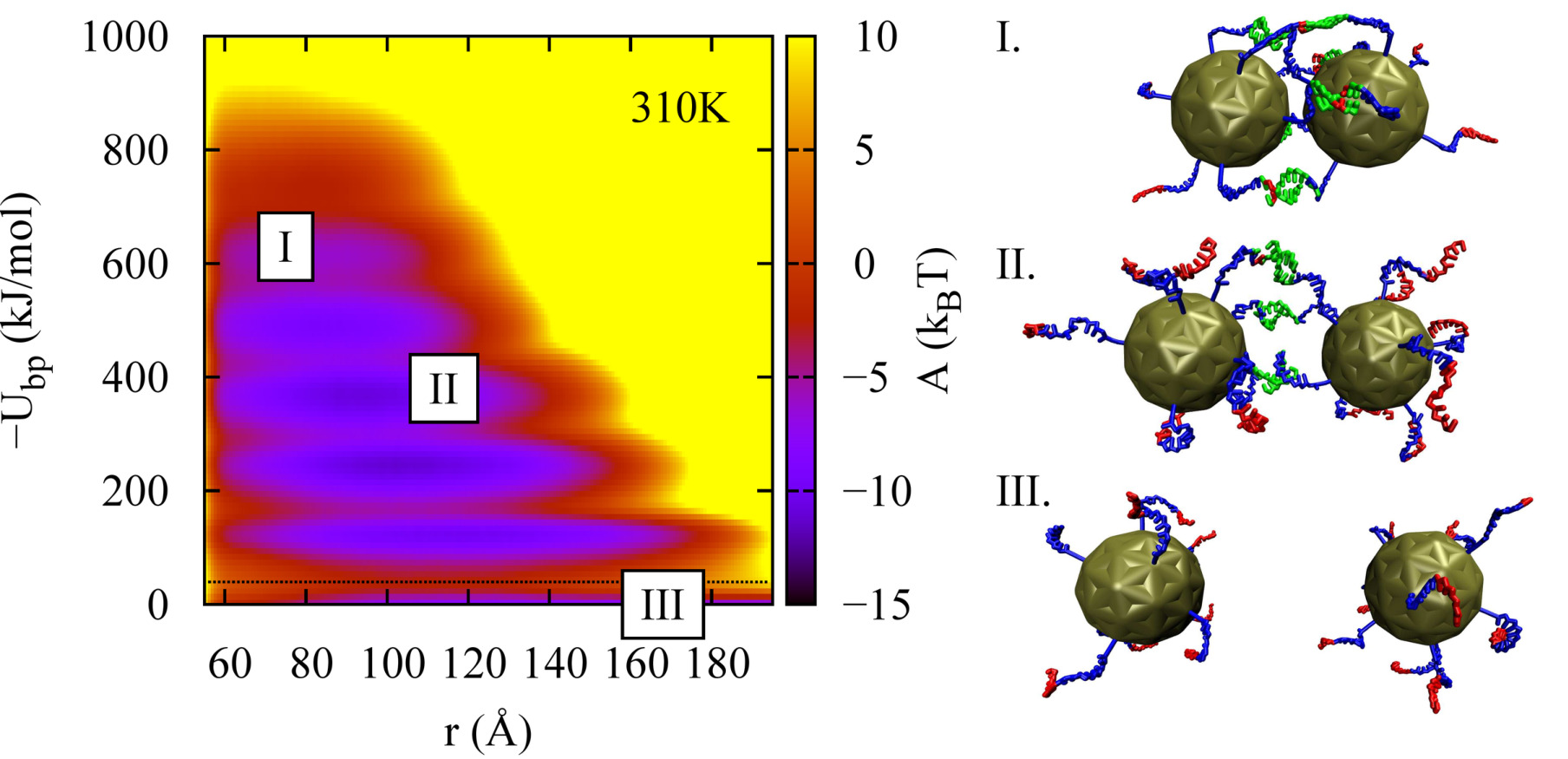
A molecular view of DNA-conjugated nanoparticle association energies
Joshua P. Lequieu, Daniel M. Hinckley, Juan J. de Pablo
Soft Matter (2015) 11:1919--1929 DOI: 10.1039/c4sm02573c
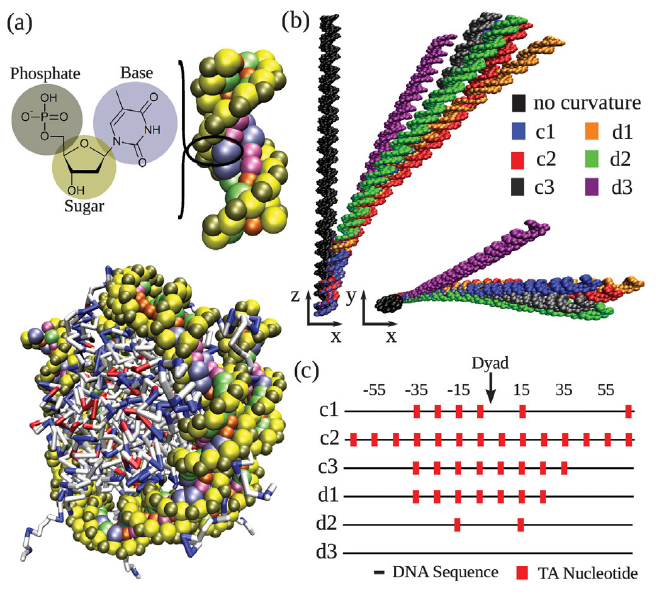
Coarse-grained modeling of DNA curvature
Gordon S. Freeman, Daniel M. Hinckley, Joshua P. Lequieu, Jonathan K. Whitmer, Juan J. de Pablo
J. Chem. Phys. (2014) 141:165103 DOI: 10.1063/1.4897649
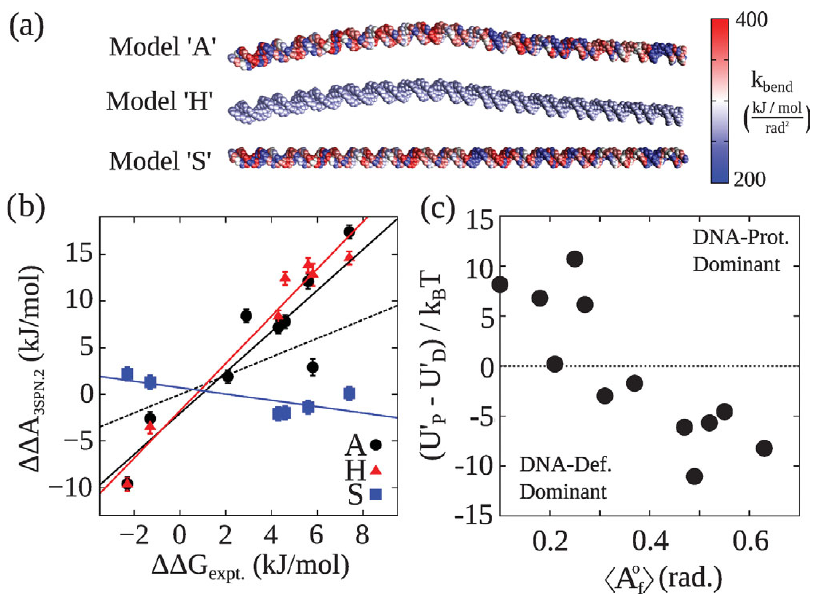
DNA shape dominates sequence affinity in nucleosome formation
Gordon S. Freeman, Joshua P. Lequieu, Daniel M. Hinckley, Jonathan K. Whitmer, Juan J. de Pablo
Phys. Rev. Lett. (2014) 113:1--19 DOI: 10.1103/PhysRevLett.113.168101
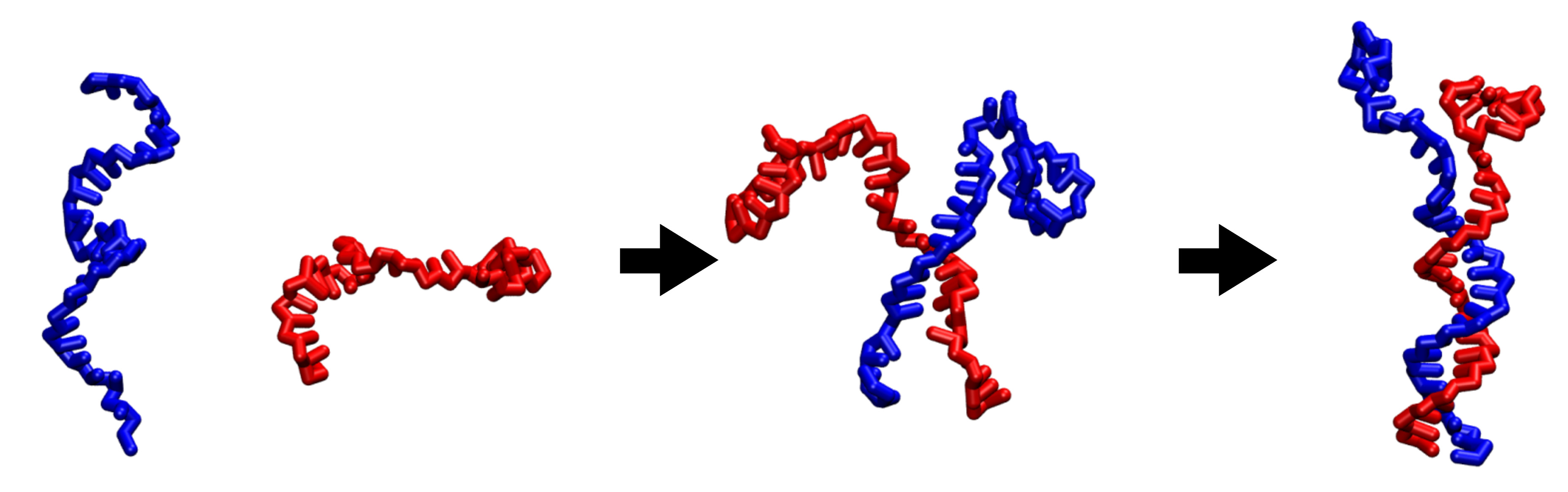
Coarse-grained modeling of DNA oligomer hybridization: Length, sequence, and salt effects
Daniel M. Hinckley, Joshua P. Lequieu, Juan J. de Pablo
J. Chem. Phys. (2014) 141:14914 DOI: 10.1063/1.4886336
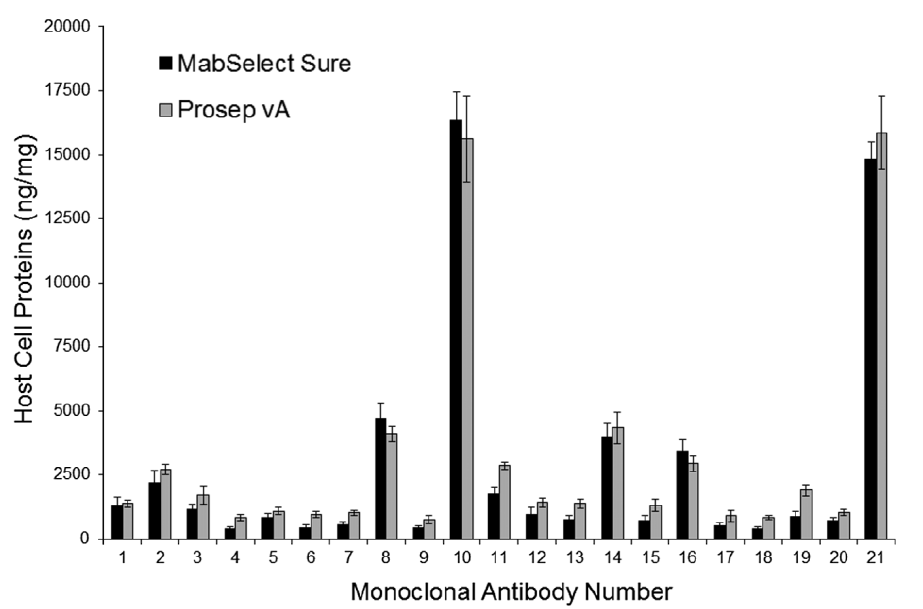
Studying host cell protein interactions with monoclonal antibodies using high throughput protein A chromatography
Vikram N. Sisodiya, Joshua Lequieu, Maricel Rodriguez, Paul McDonald, Kathlyn P. Lazzareschi
Biotechnol. J. (2012) 7:1233--1241 DOI: 10.1002/biot.201100479
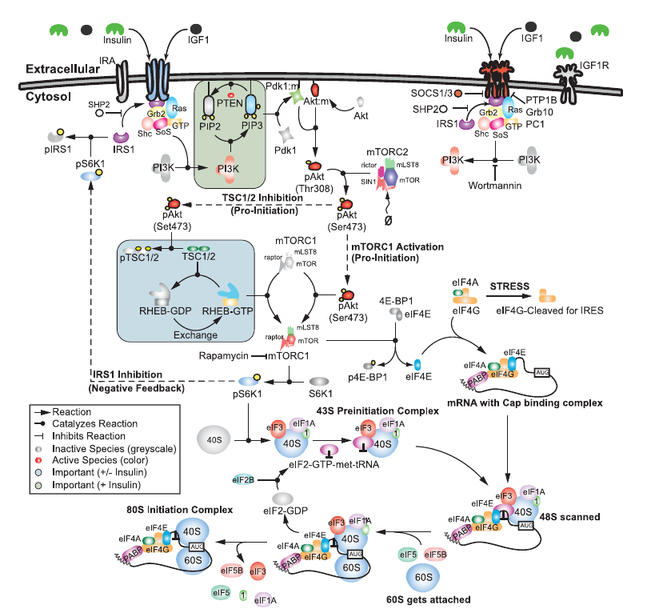
Computational modeling and analysis of insulin induced eukaryotic translation initiation
Joshua Lequieu, Anirikh Chakrabarti, Satyaprakash Nayak, Jeffrey D. Varner
PLoS Comp. Biol. (2011) 7:e1002263 DOI: 10.1371/journal.pcbi.1002263